The human genome project is an ambitious initiative aimed at sequencing every piece of human DNA. The project attracted collaborators from research institutions around the world, including the Whitehead biomedical Institute at the Massachusetts Institute of Technology (MIT), and was finally completed in 2003.
Now, more than 20 years later, MIT professor Jonathan Weissman and his colleagues have gone beyond sequences and proposed the first comprehensive functional map of genes expressed in human cells. The data of this project was published online in cell on June 9. It links each gene with its work in cells, which represents the crystallization of cooperation in the single cell sequencing method perturb SEQ over the years.
These data can be used by other scientists. Weissman said, "this is a great resource, just as the human genome is a great resource. You can go in and do discovery based research. You don't have to define the biology you want to study in advance. You have this genotype phenotype diagram. You can go in and screen the database without doing any experiments. " He is also a member of the Whitehead Institute and an investigator of the Howard Hughes Institute of medicine.
This enables researchers to study various biological problems in depth. They used it to explore the effects of genes with unknown functions on cells, study the response of mitochondria to stress, and screen out genes that cause chromosome loss or increase. This phenotype has proved difficult to study in the past. "I think this data set will enable people from other fields of biology to carry out various analyses that we haven't even thought of, and suddenly they have something to use," said Tom Norman, the co-author of this study and a former Weisman laboratory doctor.
Pioneering perturb SEQ method
The perturb SEQ method used by the project makes it possible for scientists to track the effects of turning genes on or off in unprecedented depth. This method was first published by a group of researchers including Weisman and MIT professor Aviv Regev in 2016, but it can only be used for small-scale gene sets and costs a lot.
A large number of perturb SEQ maps are contributed by the basic work of Joseph replogle, who is a medical doctor in Weissman laboratory and one of the first authors of this paper. Replogle worked with Norman, Britt Adamson (assistant professor of molecular biology at Princeton University) and a team of 10x genomics to create a new version of perturb SEQ that could be scaled up. In 2020, researchers published a paper on proof of concept in Nature Biotechnology.
The perturb SEQ method uses crispr-cas9 genome editing to introduce genetic changes into cells, and then uses single-cell RNA sequencing to capture information about RNA expressed due to specific genetic changes. Since RNA controls all aspects of cell behavior, this approach can help decipher many cellular effects of genetic changes.
Since their initial proof of concept paper, Weissman, Regev and others have used this sequencing method on a smaller scale. For example, in 2021, researchers used perturb SEQ to explore how human and viral genes interact in the process of infection with HCMV, a common herpes virus.
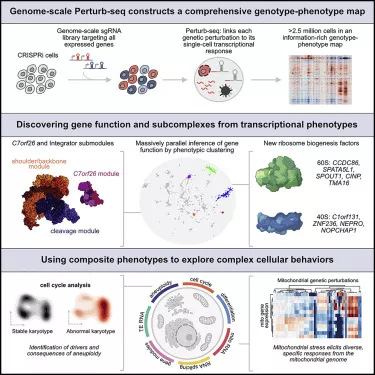
In the new study, replogle and collaborators including Weissman lab graduate student and the paper's co-author Reuben Saunders extended the method to the entire genome. Using human blood cancer cell lines and non cancerous cells from the retina, he conducted perturb SEQ on more than 2.5 million cells and used these data to build a comprehensive map linking genotypes and phenotypes.
In depth study of data
After the screening, the researchers decided to put their new data set into use and begin to study some biological problems. Tom Norman pointed out: "the advantage of perturb SEQ is that it allows you to obtain a large data set in an unbiased way. No one fully knows what the limit you can get from this data set is. Now the question is, what do you actually do with it?"
The first and most obvious application is to study genes with unknown functions. Because the screen also reads the phenotypes of many known genes, researchers can use these data to compare unknown genes with known genes and look for similar transcriptional results, which may indicate that these gene products work together as part of a larger complex.
A mutation in a gene called c7orf26 is particularly striking. The researchers noted that those genes that were removed to cause a similar phenotype were part of a protein complex called integrator that played a role in creating small RNA. The integrator complex consists of many smaller subunits -- previous studies have shown that there are 14 separate proteins -- and researchers can confirm that c7orf26 constitutes the 15th component of the complex.
They also found that these 15 subunits work together in smaller modules and perform specific functions in the integrator complex. Saunders said: "it is not clear that these different modules are so different in function without the situation of thousands of feet of tall buildings rising from the ground."
Another advantage of perturb SEQ is that because the test focuses on single cells, researchers can use these data to observe more complex phenotypes, which will become ambiguous when they are studied together with data from other cells. "We often take out all the cells whose 'gene x' has been knocked out, average them together, and then see how they change," Weissman said. "But sometimes when you knock out a gene, different cells that lose the same gene will have different behaviors, and this behavior may be ignored by the average."
The researchers found that a subset of genes, whose removal leads to different results in different cells, is responsible for chromosome segregation. Their removal causes the cell to lose a chromosome or pick up an additional chromosome, a condition known as aneuploidy. "You can't predict what the transcriptional response is when you lose this gene, because it depends on what chromosome secondary effects you gain or lose," Weissman said, "We realized that we could reverse this situation, create this complex phenotype, and look for the characteristics of chromosome gain and loss. In this way, we have completed the first genome-wide screening to find the factors required for the correct separation of DNA."
"I think aneuploidy research is the most interesting application of this data so far. It captures a phenotype that you can only read with a single cell. You can't pursue it in other ways," Norman said.
The researchers also used their data set to study how mitochondria respond to stress. Mitochondria, which evolved from free-living bacteria, carry 13 genes in their genomes. In nuclear DNA, about 1000 genes are related to mitochondrial function to some extent. Replogle said, "people have been paying attention to how nuclear DNA and mitochondrial DNA are coordinated and regulated under different cell conditions, especially when a cell is under pressure."
The researchers found that when they disturbed different mitochondrial related genes, the nuclear genome responded similarly to many different genetic changes. However, mitochondrial genomes respond much more.
Replogle pointed out: "it is still an open question why mitochondria still have their own DNA. A big revelation from our work is that one advantage of having an independent mitochondrial genome may be local or very specific genetic regulation of different stressors."
"If you have one mitochondrion destroyed and another destroyed in a different way, these mitochondria may respond differently," Weissman said.
In the future, researchers hope to use perturb SEQ for different types of cells other than the cancer cell lines they began to use. In addition, they hope to continue to explore their gene function map and hope that others can do the same. Norman said, "this is really the result of years of work by the author and other collaborators. I am really glad to see it continue to succeed and expand."