Recently, the rice molecular design technology and application innovation team of the Institute of Crop Sciences of the Chinese Academy of Agricultural Sciences cooperated with Shanghai Jiaotong University to construct a high-quality rice pan genome based on the second-generation and third-generation genome sequencing data of 111 representative rice resources, and obtained high-quality reference genomes of 9 representative rice populations, including 5 intact rice genomes.
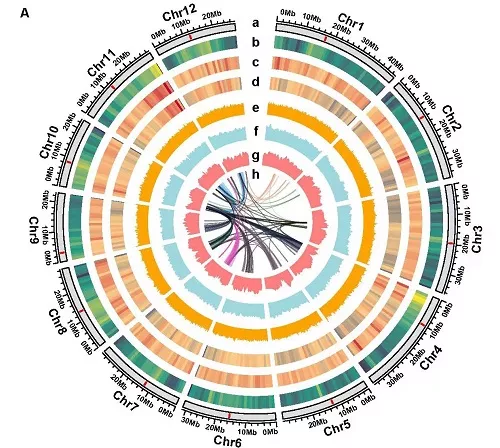
Pan genomic characteristics of 111 rice germplasm. Map provided by Chinese Academy of Agricultural Sciences
The relevant data provide an important basis for deeply mining genomic variation and excellent genes and cultivating breakthrough new rice varieties. Relevant research results are published online in genome research.
According to Wang Wensheng, a researcher at the Institute of Crop Sciences of the Chinese Academy of Agricultural Sciences, the team published a paper in nature in 2018 and conducted re sequencing and big data analysis on 3010 rice samples from 89 countries around the world. These 3010 rice samples represent about 95% of the genetic diversity of 780000 core germplasm in the world, and constructed the first complete pan genome of Asian cultivated rice. However, the previous research is based on the second-generation sequencing data. Compared with the third-generation sequencing technology, the data quality will be higher.
The pan genome constructed in this study has added three generations of sequencing data on the basis of the original. Therefore, the pan genome constructed is more accurate, continuous and complete, including 879 MB non redundant new sequences, involving 19319 new protein coding genes (2132 new gene families).
Compared with the second-generation sequencing data, the third-generation sequencing data has a lower false positive rate in detecting gene presence deletion variation (PAV), especially for genes containing repetitive sequences. In addition, 14471 presence deletion variants were significantly associated with multiple agronomic traits, indicating that gene presence deletion variants may make an important contribution to rice phenotypic variation.
The high-quality rice genome, pan genome, gene existence deletion variation and other resources obtained in this study are conducive to promoting the research of rice functional genome, deeply excavating genome variation and excellent genes, and providing an important basis for cultivating breakthrough new rice varieties.
The research was supported by the National Natural Science Foundation of China, the project of Hainan yazhouwan seed laboratory, the scientific and technological innovation project of the Chinese Academy of Agricultural Sciences and the national high-level talent support plan.
Relevant paper information:< https://doi.org/10.1101/gr.276015.121 >